Tutorial 02: How to upload your own data
In this tutorial we will show you how to upload your own dataset. We begin in to the datasets menu.
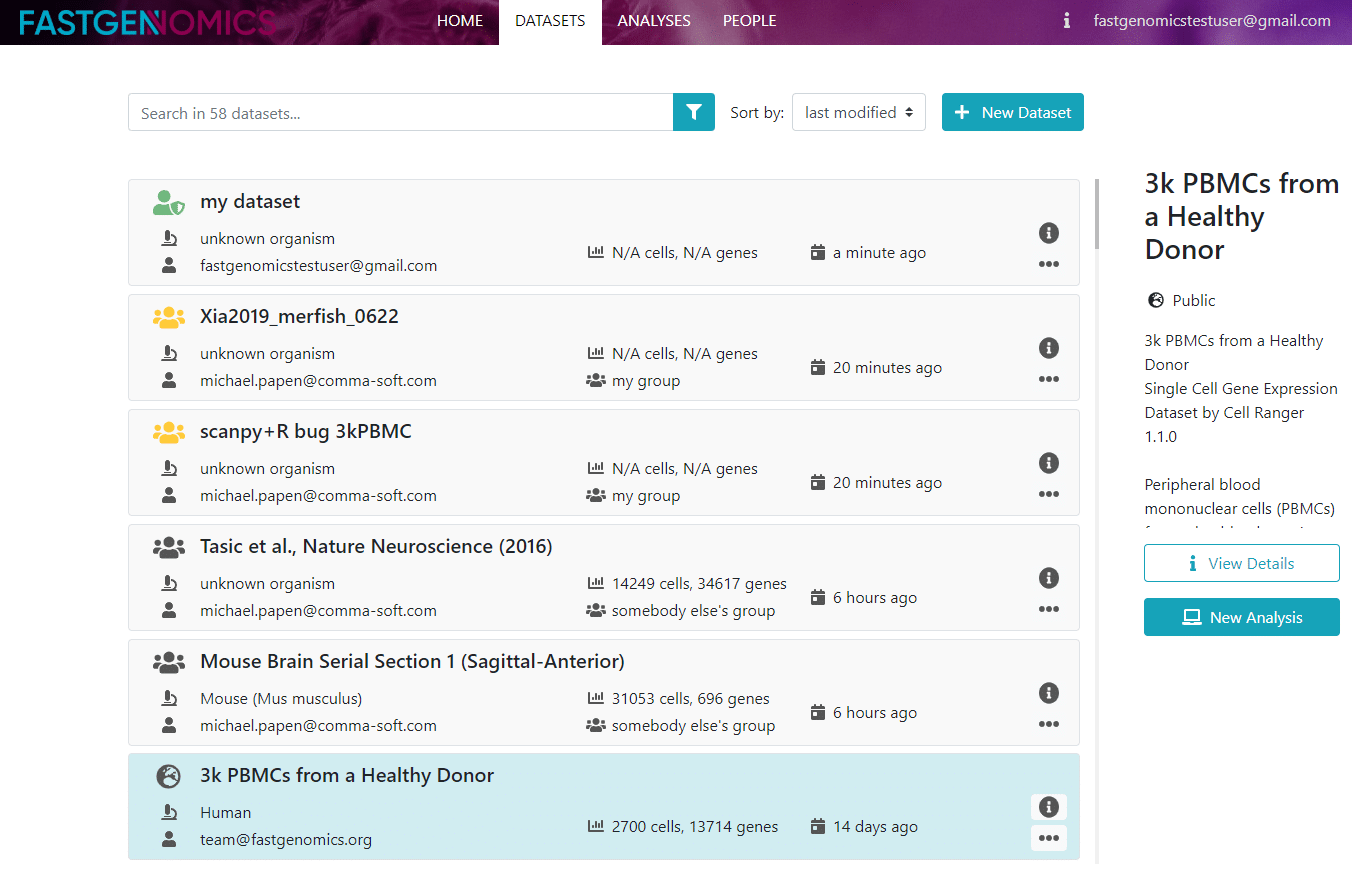
After we click on the plus sign in the top right corner, the data upload process can begin. A new window pops up, in which you need to specify the details of your data.
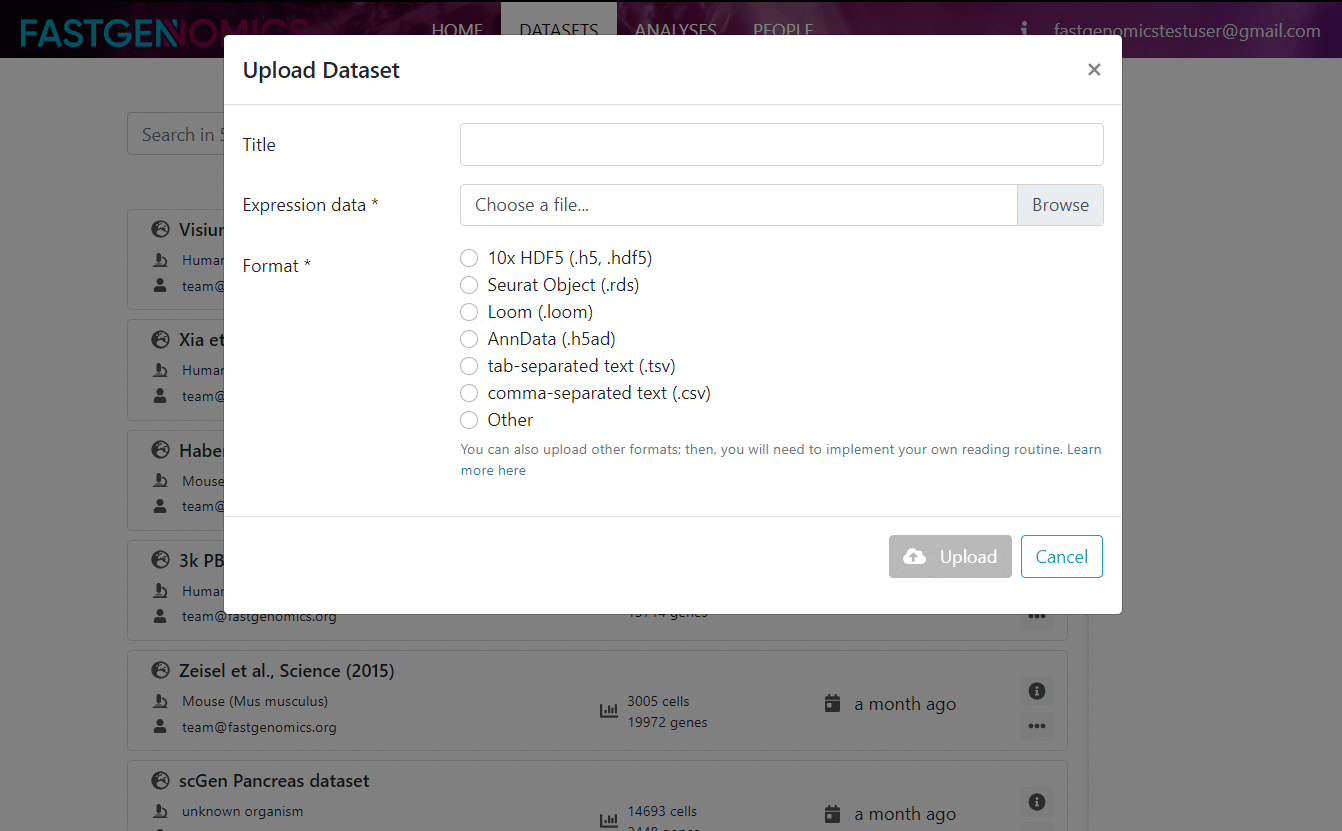
The information you need to fill in at first are the title of the dataset and the file format. Then you can choose the file on your local hard drive that shall be uploaded to FASTGenomics. In the upload dialog, you can see the currently supported file formats on FASTGenomics, namely
- 10x (*.hdf5)
- Seurat (*.rds)
- Anndata (*.h5ad)
- Loom (*.loom)
- Tab-/Comma-separated text (*.tsv, *.csv)
- Other (it is possible to implement your own loading routine)
When you click on upload, the upload process begins and you are redirected to the metadata page. Here, you can enter many more details such as a free description of your dataset (where you can use html to structure your text if you like), your contact details, tissue and technology information and additional information such as a citation and data repository. If your data is preprocessed, you can select the current normalization of your data, for example counts, logarithmic counts, or others.
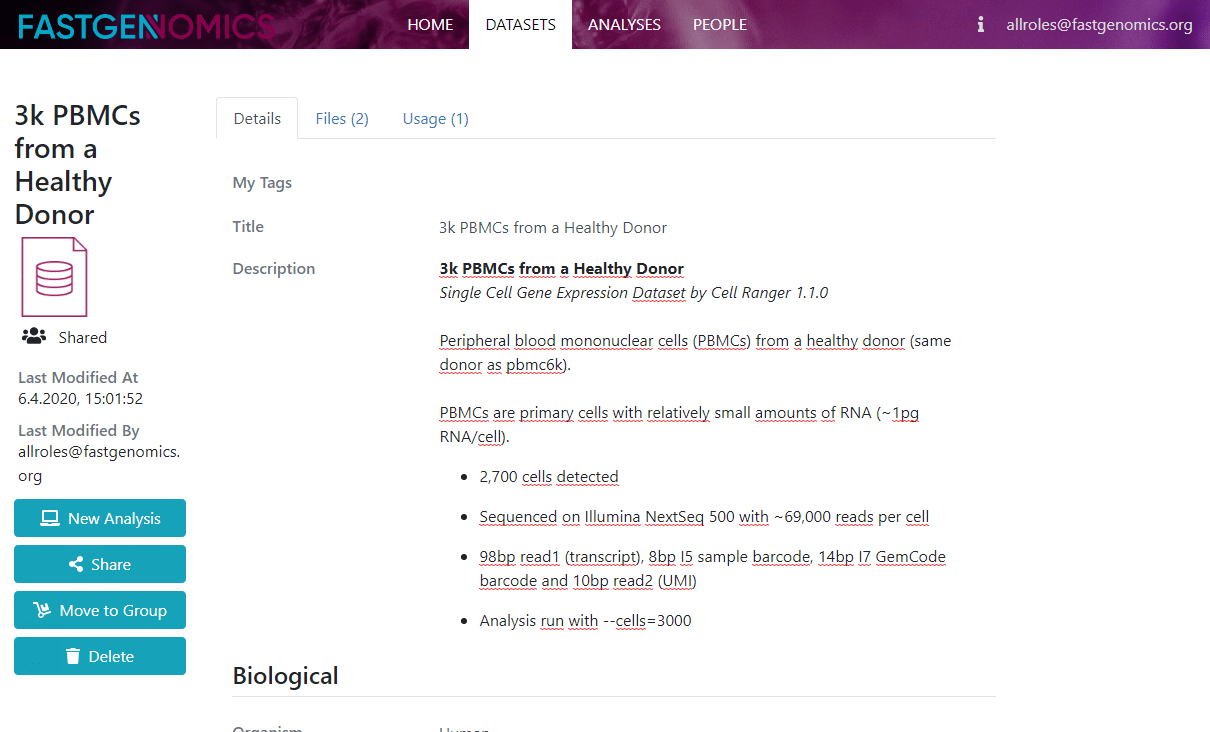
Note that we store the complete file and not just the expression matrix. For certain formats, such as Anndata, this means that you can also pass already computed layers to the FASTGenomics database and make much more available than just the raw data. If you do so, please write about the involved processing in teh description field.
When the upload process is finished and you have completed adding metadata, you might want to share the dataset with your colleagues. FASTGenomics makes this very easy: just click on the share button in the dataset menu (after you highlighted the corresponding dataset) or on the dataset details page. A share dialog will pop up and will ask you with whom you would like to share the dataset. Just enter the email of your colleague and select to share. Note that in order to see your colleague, she of he needs to Allow other users to find me. This option can be checked in your profile settings.
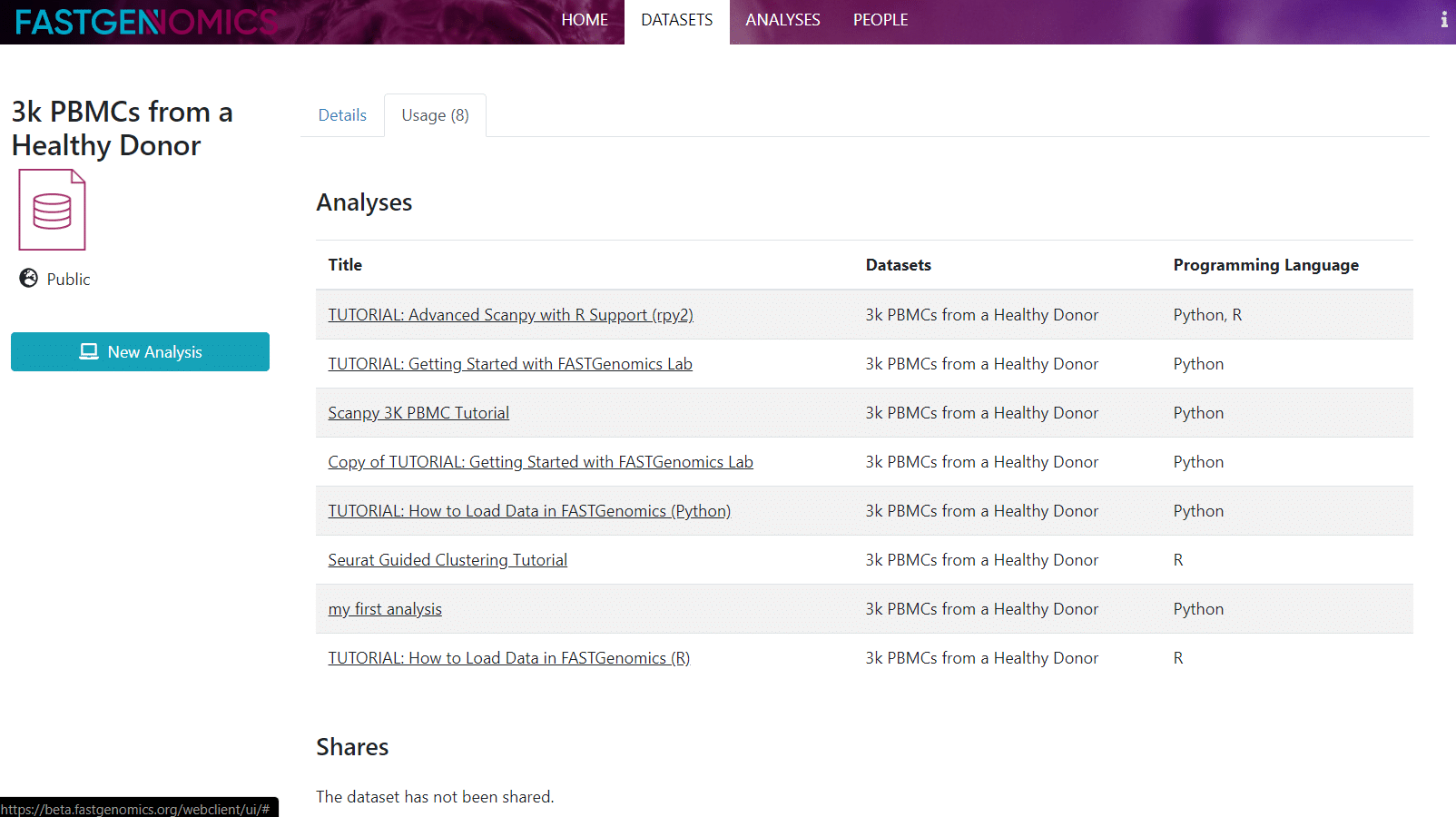
If you shared your dataset already, you can click on the usage tab at the top to check with whom you shared it (in case you forgot) and also in which analyses your dataset is used.
This concludes our short tutorial on datasets. If you have further questions, please get in contact.
